Annotate genetic variants by m6A function
Characterize genetic variants that specially alter m6A in RNA molecules
Robust prediction model (AUC>85%)
Statistics, analysis and visualization
Get started!
Characterize genetic variants that specially alter m6A in RNA molecules
Robust prediction model (AUC>85%)
Statistics, analysis and visualization

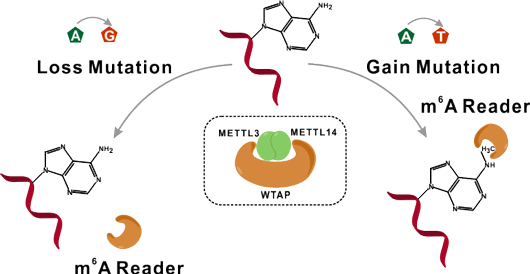
N6-methyladenosine (m6A) is one of the most abundant modifications in RNA molecules, and regulates almost all aspects of RNA metabolism. Genetic variants especially cancer mutations can significantly alter the status of m6A, leading to the gain or loss of N6-methyladenosine. m6ASNP is specifically designed to characterize such genetic mutations and potential roles of m6A alterations may be derived from our prediction tool.
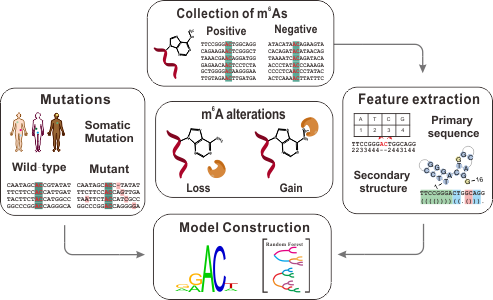
To construct the prediction model, we extracted 60 sequence distribution and 27 secondary structure features from the flanking regions around the methylated adenosine. With the Random Forest algorithm, we have trained two accurate prediction models for human and mouse, respectively. The evaluation on additional test data set has proven that the area under ROC curve (ROC) of both models are larger than 85%.
 Baker, Paul T. et al. Phys.Rev 2015
Baker, Paul T. et al. Phys.Rev 2015
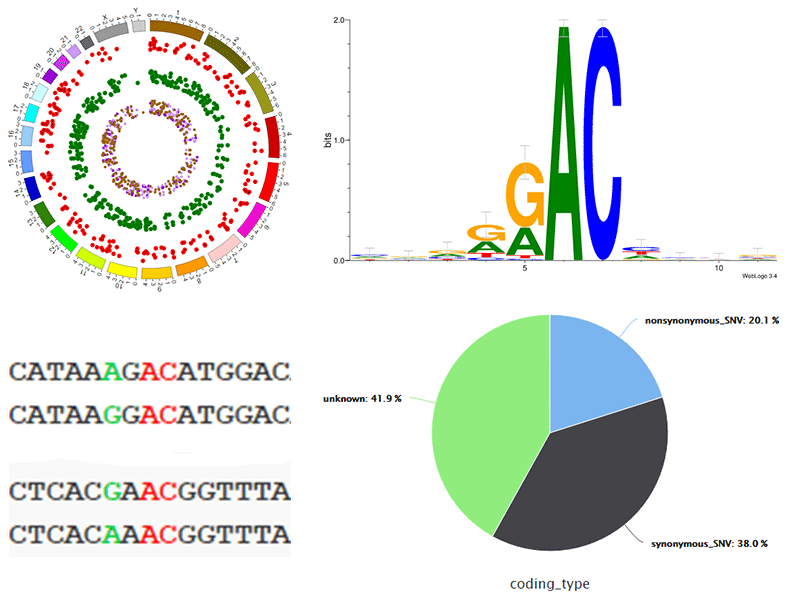
The web server will conduct a comprehensive annotation and statistical analysis for all the predicted m6A alterations. Serval statistical diagrams and a genome browser are provided to visualize the topology characteristics of predicted m6A alterations.